Fitting single crystal parameters with DAMASK simulations#
This notebook processes the results from the demo workflow fit_single_crystal_parameters, and generates a plot of stress strain both simulated and experimental, showing the model converging on the desired deformation behavior with each pertubation of the plastic single crystal parameters.
import matflow as mf
import matplotlib.pyplot as plt
import numpy as np
from pathlib import Path
Utility functions#
def clean_nonetypes(stress_array, strain_array):
"""
For some reason experimental data has nonetypes:
This function cleans the stress array of nonetypes
and also ensures the strain array is matched to it.
"""
# DEBUG
# print(f"input array : {stress_array}")
# print(f"bool array of nonetypes : {stress_array != np.array(None)}")
cleaned_stress_array = stress_array[stress_array != np.array(None)]
cleaned_strain_array = strain_array[stress_array != np.array(None)]
return cleaned_stress_array, cleaned_strain_array
Define workflow#
workflow_path = Path("fit_single_crystal_parameters_2025-05-28_143539")
workflow = mf.Workflow(workflow_path)
Your workflow output (from matflow show -f) should look like this:
┌────┬───────────────────────────────────────────────┬──────────┬────────────────────────────┬────────────────────────────────────────────────┐
│ ID │ Name │ Status │ Times │ Actions │
├────┼───────────────────────────────────────────────┼──────────┼────────────────────────────┼────────────────────────────────────────────────┤
│ 33 │ fit_single_crystal_parameters_2025-05-28_143… │ inactive │ sb. 2025-05-28 14:35:41 │ generate_microstructu… 0 | ■ │
│ │ │ │ st. 16:36:35 │ generate_volume_eleme… 0 | ■ │
│ │ │ │ en. 16:36:31 │ read_tensile_test_fro… 0 | ■ │
│ │ │ │ │ simulate_VE_loading_d… 0 | ■■■■■■■■■■■■■■ │
│ │ │ │ │ 1 | ■■■■■■■■■■■■■■ │
│ │ │ │ │ 2 | ■■■■■■■■■■■■■■ │
│ │ │ │ │ 3 | ■■■■■■■■■■■■■■ │
│ │ │ │ │ fit_single_crystal_pa… 0 | ■■
└────┴───────────────────────────────────────────────┴──────────┴────────────────────────────┴────────────────────────────────────────────────┘
Extract and clean data#
# show attrs of workflow obj with dir:
dir(workflow)
['_IndexPath1',
'_IndexPath2',
'_IndexPath3',
'_Workflow__EAR_obj_map',
'_Workflow__template_components',
'_Workflow__wait_for_direct_jobscripts',
'_Workflow__wait_for_scheduled_jobscripts',
'__annotations__',
'__class__',
'__delattr__',
'__dict__',
'__dir__',
'__doc__',
'__eq__',
'__format__',
'__ge__',
'__getattribute__',
'__gt__',
'__hash__',
'__init__',
'__init_subclass__',
'__le__',
'__lt__',
'__module__',
'__ne__',
'__new__',
'__reduce__',
'__reduce_ex__',
'__repr__',
'__setattr__',
'__sizeof__',
'__slots__',
'__str__',
'__subclasshook__',
'__weakref__',
'_abort_run',
'_accept_pending',
'_add_empty_loop',
'_add_empty_task',
'_add_file',
'_add_loop',
'_add_parameter_data',
'_add_submission',
'_add_task',
'_add_unset_parameter_data',
'_app',
'_app_attr',
'_check_loop_termination',
'_creation_info',
'_default_ts_fmt',
'_default_ts_name_fmt',
'_delete_no_confirm',
'_exec_dir_name',
'_get_empty_pending',
'_get_new_task_unique_name',
'_in_batch_mode',
'_input_files_dir_name',
'_is_tracking_unset',
'_loops',
'_merged_parameters_cache',
'_name',
'_pending',
'_reject_pending',
'_reset_pending',
'_resolve_input_source_task_reference',
'_resolve_singular_jobscripts',
'_set_file',
'_store',
'_submissions',
'_submit',
'_tasks',
'_template',
'_template_components',
'_tracked_unset',
'_ts_fmt',
'_ts_name_fmt',
'_use_merged_parameters_cache',
'_write_empty_workflow',
'abort_run',
'add_loop',
'add_submission',
'add_task',
'add_task_after',
'add_task_before',
'artifacts_path',
'batch_update',
'cached_merged_parameters',
'cancel',
'check_parameters_exist',
'copy',
'creation_info',
'delete',
'elements',
'ensure_commands_file',
'execute_combined_runs',
'execute_run',
'execution_path',
'from_JSON_file',
'from_JSON_string',
'from_YAML_file',
'from_YAML_string',
'from_file',
'from_template',
'from_template_data',
'get_EAR_IDs_of_tasks',
'get_EAR_skipped',
'get_EARs_from_IDs',
'get_EARs_of_tasks',
'get_all_EARs',
'get_all_element_iterations',
'get_all_elements',
'get_all_parameter_data',
'get_all_parameter_sources',
'get_all_parameters',
'get_all_submission_run_IDs',
'get_element_IDs_from_EAR_IDs',
'get_element_iteration_IDs_from_EAR_IDs',
'get_element_iterations_from_IDs',
'get_element_iterations_of_tasks',
'get_elements_from_IDs',
'get_iteration_task_pathway',
'get_parameter',
'get_parameter_data',
'get_parameter_set_statuses',
'get_parameter_source',
'get_parameter_sources',
'get_parameters',
'get_process_IDs',
'get_run_directories',
'get_running_elements',
'get_running_runs',
'get_scheduler_job_IDs',
'get_store_EARs',
'get_store_element_iterations',
'get_store_elements',
'get_store_tasks',
'get_task_IDs_from_element_IDs',
'get_task_elements',
'get_task_unique_names',
'get_text_file',
'id_',
'input_files_path',
'is_parameter_set',
'list_jobscripts',
'list_task_jobscripts',
'loops',
'name',
'num_EARs',
'num_added_tasks',
'num_element_iterations',
'num_elements',
'num_loops',
'num_submissions',
'num_tasks',
'path',
'process_shell_parameter_output',
'rechunk',
'rechunk_parameter_base',
'rechunk_runs',
'reload',
'resolve_jobscripts',
'save_parameter',
'set_EAR_end',
'set_EAR_skip',
'set_EAR_start',
'set_EARs_initialised',
'set_multi_run_ends',
'set_multi_run_starts',
'set_parameter_value',
'set_parameter_values',
'show_all_EAR_statuses',
'store_format',
'submissions',
'submissions_path',
'submit',
'task_artifacts_path',
'tasks',
'template',
'template_components',
'temporary_rename',
'ts_fmt',
'ts_name_fmt',
'unzip',
'url',
'wait',
'zip']
## Experimental Stress-strains ##
exp_data = workflow.tasks.read_tensile_test_from_CSV.elements[0].iterations[0].outputs.tensile_test.value
exp_stress = np.array(exp_data['true_stress'])
exp_strain = np.array(exp_data['true_strain'])
# clean experimental data of nonetypes
exp_stress, exp_strain = clean_nonetypes(exp_stress, exp_strain)
## Simulated stress-strains ##
# Getting an error? try running twice.
VE_response = workflow.tasks.simulate_VE_loading_damask.elements[0].iterations[0].outputs.VE_response.value
sim_stress = np.array(VE_response['volume_data']['vol_avg_stress']['data'])
sim_strain = np.array(VE_response['volume_data']['vol_avg_strain']['data'])
input array : [None None -34700.0 ... None None None]
bool array of nonetypes : [False False True ... False False False]
(171, 3, 3)
(171, 3, 3)
Plot simulated stress strains against experimental#
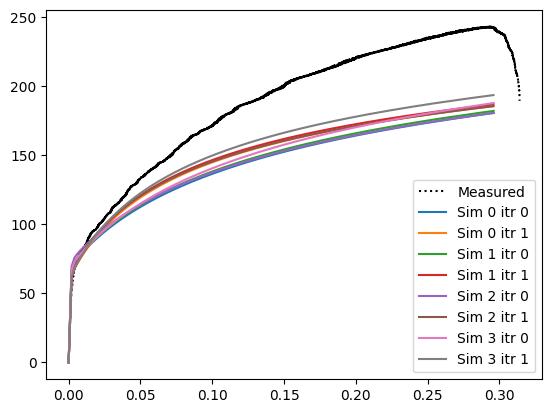
It is shown that with successive pertubations of the single crystal parameters h_0_sl-sl, xi_0_sl, and xi_inf_sl, the predicted stress-strain curve approaches the measured stress-strain curve. With the simulation more and more closely capturing the deformation behavior of the real material:
plt.figure(1)
plt.plot(exp_strain, exp_stress/1e6,
color='k',
linestyle="dotted",
label="Measured")
for e, sim in enumerate(workflow.tasks.simulate_VE_loading_damask.elements):
for i, itr in enumerate(sim.iterations):
print(f"Sim {e} iter {i}... ")
VE_response = itr.outputs.VE_response.value
sim_stress = np.array(VE_response['volume_data']['vol_avg_stress']['data'])
sim_strain = np.array(VE_response['volume_data']['vol_avg_strain']['data'])
plt.plot(sim_strain[:,0,0], sim_stress[:,0,0]/1e6,
linestyle="solid",
label=f"Sim {e} itr {i}")
plt.legend()
Sim 0 iter 0...
Sim 0 iter 1...
Sim 1 iter 0...
Sim 1 iter 1...
Sim 2 iter 0...
Sim 2 iter 1...
Sim 3 iter 0...
Sim 3 iter 1...
<matplotlib.legend.Legend at 0x150eb7443610>